DNA Library
Designed for identification of new actives against proteins essential for DNA stability
5 530 compounds
Cell cycle is a complex and multi-level process controlled by genome. Maintenance of genomic stability relies on the coordinated action of a network of cellular processes, including DNA replication, DNA repair, cell-cycle progression, and others. where DNA replication plays a pivotal role. DNA breaks can be influenced by both exogenous and/or endogenous factors. Evidently, that late detection and correction of DNA damages dramatically increase the probability of cell death. But at the same time, changes in the behavior of proteins engaged in the DNA reparation process could also cause damages, for example implementation of inappropriate bases into the chain of DNA.
Library Design
Our library comprises compounds with in silico predicted activity against key housekeeping proteins essential for DNA stability. All the targets were analyzed based on their therapeutic influence and were divided into:
- Oxidative DNA damage triggers, like Poly(ADP-ribose)polymerases (PARPs). PARP1, translates the occurrence of DNA breaks detected by zinc-finger domain into a signal, and initiates an immediate cellular response to metabolic, chemical, or radiation-induced single-strand DNA breaks. The telomere protein Tankyrase 1 regulates DNA damage responses at telomeres.
- DNA damage-stimulated kinases, like DNA-dependent protein kinase (DNA-PK), ATM and ATR kinases, Checkpoint kinases (CHK1/2). Both kinases are important for homologous recombination repair of DNA double strand breaks.
Structure-based approach was mainly used in the library design. In the cases of known crystal structures with co-crystallized inhibitors the protocol included structure alignment and common pharmacophore identification, protein site mapping and constraints assignment, docking. In case of unliganded protein structure, identification of putative binding site and key residues were performed with PocketFinder and subsequent InducedFit technique, which assumes flexible site environment.
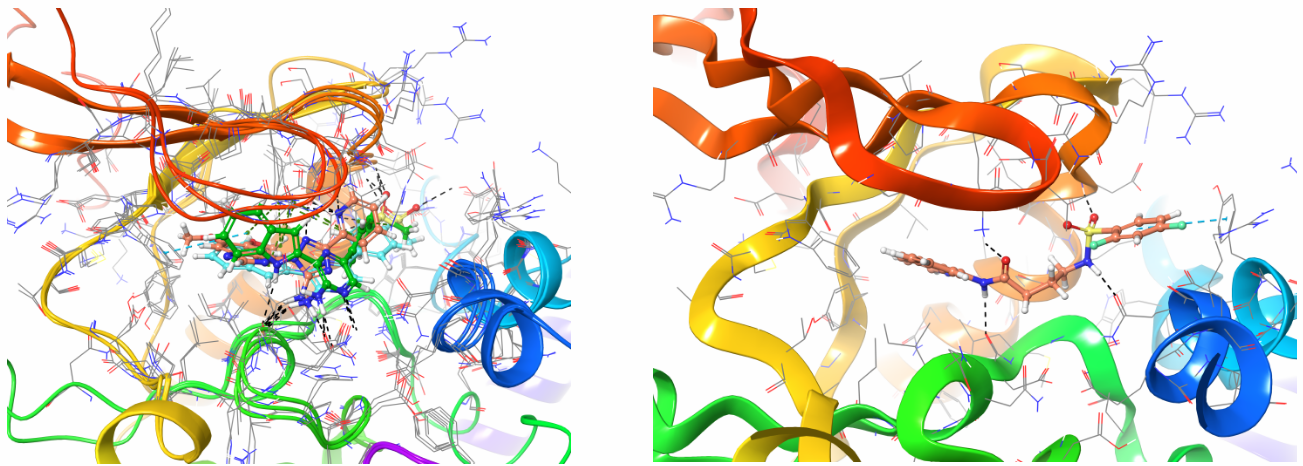
Example of InducedFit docking of the reference set from CHEMBLdb against ATM target (on the left) and Z52128042 in the same binding site after generation of constraints from key interactions and docking of entire Enamine screening database (on the right).



