Tubulin Library
Library of potential tubulins ligands
3 452 compounds
Tubulin targeted library has been designed with complex approach, including molecular docking, 2D similarity & topological analogues search, molecular and structural filters.
Library Design
Molecular docking
Protein structures recorded recently in PDB were considered and analyzed to be most suitable for in silico screening: 4YJ2, 5C8Y, 5CA1. The main features of protein-ligand interaction in the binding site of all three tubulins structures are very similar and could be superimposed. Hence, protein docking model was build based on the features of key amino acid residues in the binding pocket and ligands interactions observed in analyzed protein structures.
Docking model has been validated with set of known actives (110 ligands) and non active molecules. Calculation constraint were corrected in accordance to activity data of the reference compound set.
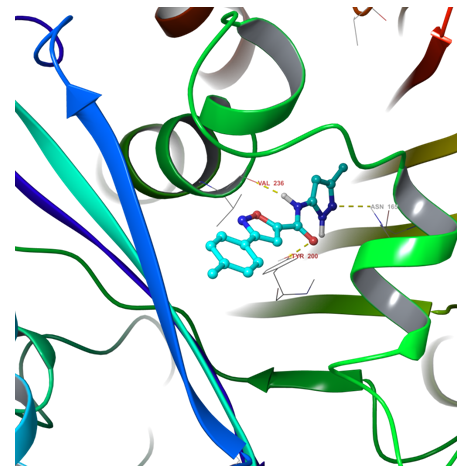
Fig. 1. Example of hit binding conformation after docking calculation.
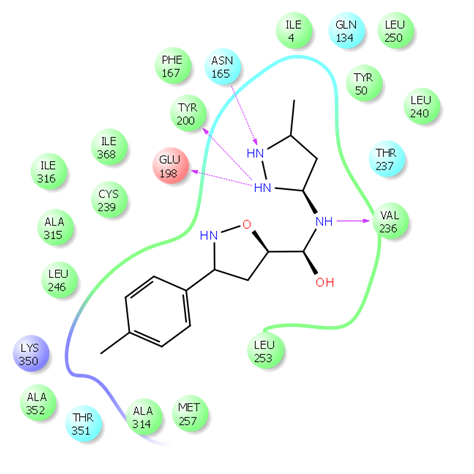
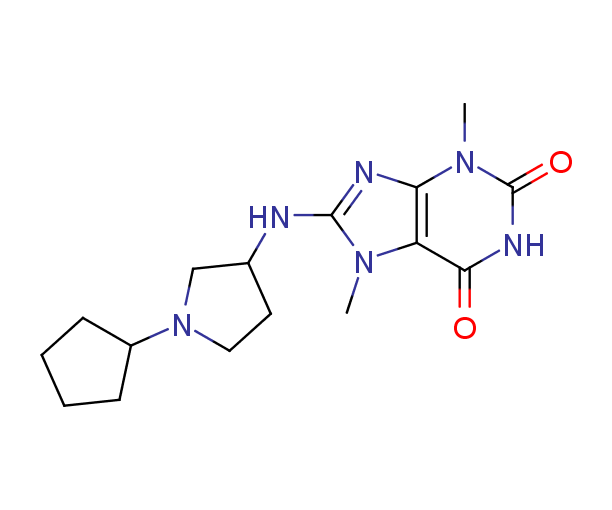
Fig. 2. 2D ligand interaction diagram of hit compound Z666232790 with high docking score.
2D Similarity search & topological analogues search
- The reference compound set was carefully compiled from available literature sources and edatabases: ChEMBL, BindingDB, PudChem.
- Tanimoto similarity range of 95–80% was used for compounds selection.
- Topological fields and bioisosteric group replacement were used to search for analogues of the most potent tubulin inhibitors.
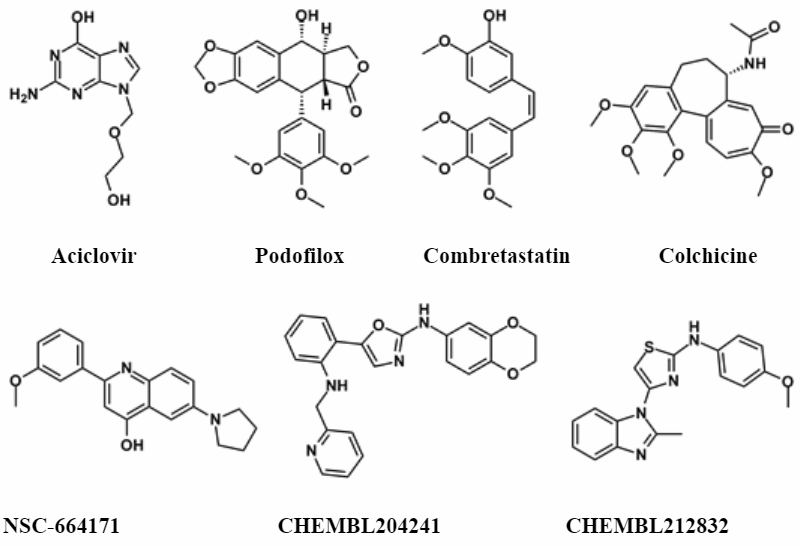
Approximately 900 compounds were selected using this approach.
Filters applied to the Library:
- In-house developed MedChem filters of unwanted structural fragment were applied to Enamine stock compound Collection as pre-selection procedure;
- PAINS, Eli Lilly, REOS and trivial functionalities filters included;
- Full Rule of Five compliance.